We are in the process of overhauling our site to make it easier to access all the information about the Data Exchange.
However, the sections below should provide an overview of the activities and resources available now.
This site is dedicated to providing updates on developments and documentation on the Data Exchange platform, data models and tools produced by the ClinGen Data Exchange Working Group.
The ClinGen Data Exchange is a comprised of the platform, data models and tools that enable an environment of standardized exchange of genomic knowledge. The Data Exchange is designed to enable a consistent approach to produce and consume the full suite of genomic knowledge curated in ClinGen. Access to the Data Exchange is currently accessible to all systems within ClinGen and may potentially be made available to external systems in the future.
Collaborations, Interactions & Integrations
- Allele Registry - The ClinGen Allele Registry has adopted the Allele Model.
- VCI's ClinVar Submission Process - Tools have been developed to transform and generate the internal VCI representation of variant interpretations into valid 'ClinVar' submission records. Currently manual, soon to be integrated with the VCI's User Interface.
- SEPIO - The Monarch Intiative's Scientific Evidence Provenance Information Ontology has become the basis for the Interpretation modeling approach. Plans are to continue developing all future knowledge models using the approach developed here.
- GA4GH - ClinGen is a driver project for the GA4GH and heavily involved in the Genomic Knowledge Standards (GSK) workstream to harmonize our models for both Variants (aka Alleles) and Variant Annotations (aka Interpretations). The ClinVar Submitter tools are planned to be a pilot project to illustrate the utility of the GA4GH GKS Variant model.
- HL7 Clinical Genomics(CG) - ClinGen data exchange modeling members actively participate in HL7's CG WG to provide a communication channel for overlapping objectives.
Data Models
- The Allele Model was released in 2017 and is the basis for the JSON-LD representation of alleles in the ClinGen Allele Registry.
- The Interpretation Model was released in June 2018. This first interpertation model was scoped to provide support for Variant Pathogenicity Interpretations as specified by the 2015 ACMG ISV guidelines.
Current Plans
There will be several more Interpretation models developed and implemented to support the sharing of the other types of curated genomic knowledge within ClinGen in the upcoming months. These models will be documented on this site once released.
Platform
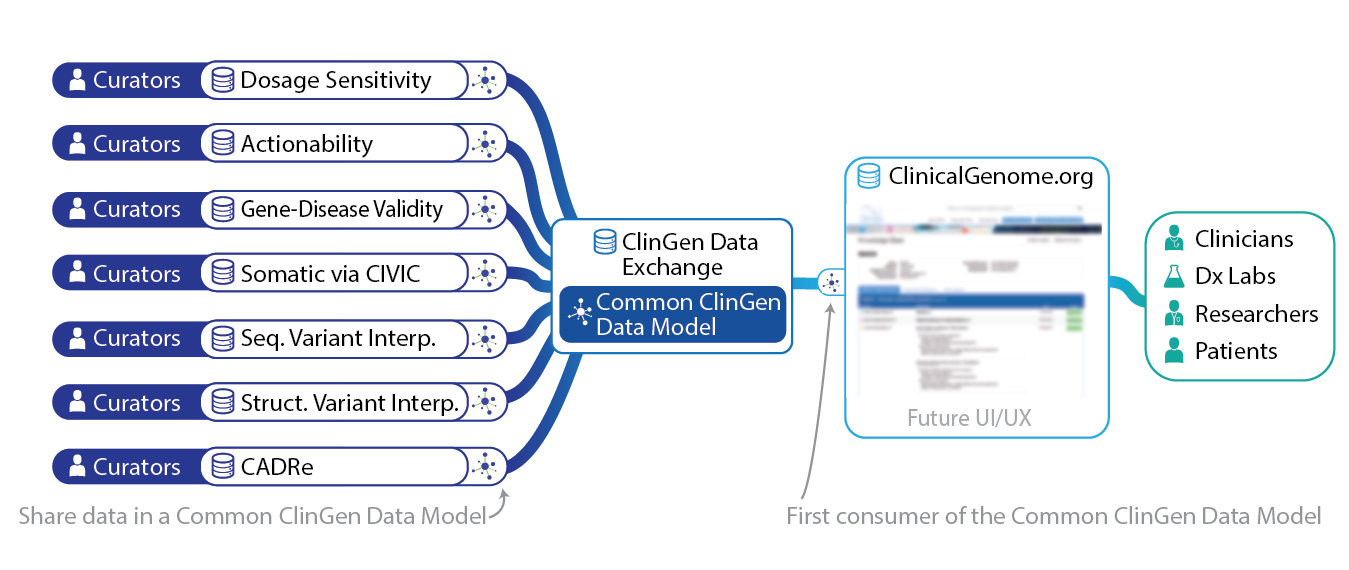
The figure below illustrates the current platform also referred to as the ClinGen Data Exchange. Currently, the primary consumer of the platform data is the ClinGen website. And, several of the ClinGen curation systems are actively producing and sharing their data with the platform to be accessible on the website.
In the upcoming phase of platform development all curated genomic knowledge types will be modified to standardized models by the data exchange working group. And further integration opportunities outside the website will be explored.
Tools
Information on the tools available for working with our models is currently located on the Tools page in the Interpretation Model. This may be re-organized and moved in the future. This section will provide updates on how to access the information.